|
Back to Main Research page |
|
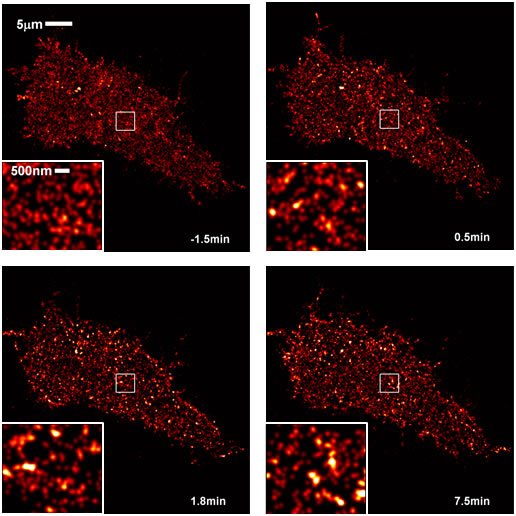
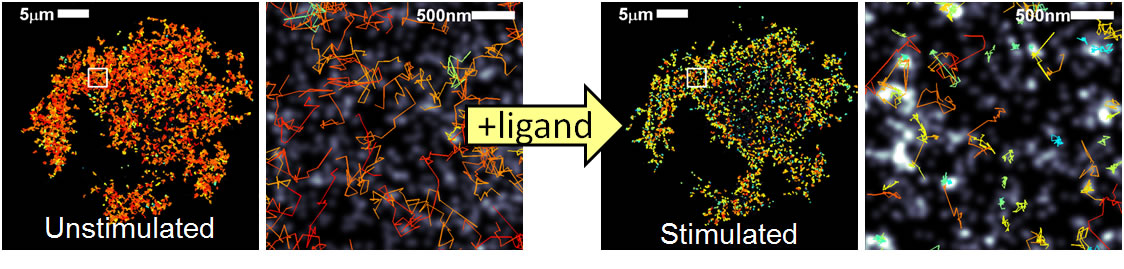
Super-Resolution Imaging of IgE Receptor SignalingUsing super-resolution stochastic optical reconstruction microscopy (STORM), we image proteins on the plasma membrane of live RBL-2H3 mast cells with ~25nm resolution. We have quantitatively mapped the rearrangement of IgE receptors in response to receptor cross-linking caused by the binding of trivalent ligands (Fig 1). These ligands are based on double-stranded DNA , and their structures are precisely defined. Live cell STORM is also being used to track single receptor trajectories and study protein diffusion dynamics as ligand stimulation proceeds (Fig 2). Ongoing experiments are aimed at quantifying the ligand stimulation-dependent co-redistribution of IgE receptors with early signaling partners using multi-color STORM. Our ultimate goal is to uncover the physical mechanisms behind the early stages of signaling, including contributions from ligand structure, membrane phase heterogeneity, protein-protein interactions, and the cytoskeleton. Shelby, S.A., D. Holowka, B. Baird and S.L. Veatch: Distinct Stages of Stimulated FceRI Receptor Clustering and Immobilization Are Identified through Superresolution Imaging. Biophys. J. 105(10): 2343-2354 (2013). Shelby, S.A., S.L. Veatch, D.A. Holowka and B.A. Baird: Functional nanoscale coupling of Lyn kinase with IgE-FceRI is restricted by the actin cytoskeleton in early antigen-stimulated signaling. Mol. Biol. Cell 27(22): 3645-3658 (2016). |
|
|
Fig 1. STORM images of IgE/FceRI on cells stimulated with trivalent ligand at 0 min. IgE/FceRI become more clustered with time after stimulation |
|
 |
|
| Fig 2. STORM images of Alexa647-IgE/FceRI acquired before (left) and after (right) ligand stimulation overlaid with receptor trajectories for the same time periods show slowed diffusion after stimulation. | |
Investigating EGF receptor signaling with micropatterned surfacesWe are investigating the spatial and temporal organization of epidermal growth factor (EGF) receptor signaling. Previous work in our laboratory provided evidence that phosphoinositide synthesis and the actin cytoskeleton contribute to stabilization of EGF receptor signaling complexes concentrated at micron-sized EGF-patterned surfaces. However, limited information exists on the structural basis for the formation of these complexes, as well as the sequelae of protein recruiting events to these receptor complexes. Building on previous work in our laboratory, we are preparing patterned ligand arrays: spatially defined micron- and submicron-sized features of EGF covalently attached to glass substrates. This unique platform enables EGF receptor activities to be imaged in fixed or live cells with high-resolution, two-color, total internal reflection fluorescence microscopy (TIRFM). We use correlation coefficient analysis to quantify the extent of protein colocalization and recruitment to the patterned EGF. Current experiments are aimed at further characterization of the spatiotemporal aspects of cell signaling by this receptor, its downstream effector Ras, and the actin cytoskeleton. Singhai, A., D.L. Wakefield, K.L. Bryant, S.R. Hammes, D. Holowka and B. Baird: Spatially Defined EGF Receptor Activation Reveals an F-Actin-Dependent Phospho-Erk Signaling Complex. Biophys. J. 107(11): 2639-2651 (2014). |
|
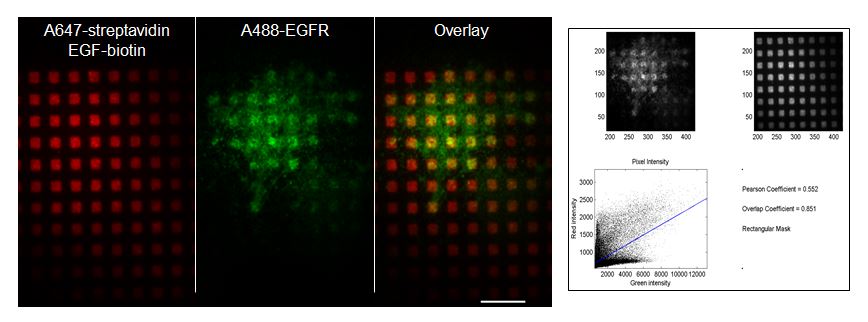 NIH-3T3 cells plated on EGF-presenting patterns (red) display colocalization of antibody-labeled EGF receptor (green) over ~2 µm square features as imaged with TIRFM (Scale bar: 10 µm). The degree of colocalization is evaluated and measured by plotting the distribution of pixel intensities from each channel and calculating aPearson’s correlation coefficient as well as an Overlap coefficient. NIH-3T3 cells plated on EGF-presenting patterns (red) display colocalization of antibody-labeled EGF receptor (green) over ~2 µm square features as imaged with TIRFM (Scale bar: 10 µm). The degree of colocalization is evaluated and measured by plotting the distribution of pixel intensities from each channel and calculating aPearson’s correlation coefficient as well as an Overlap coefficient. |
|
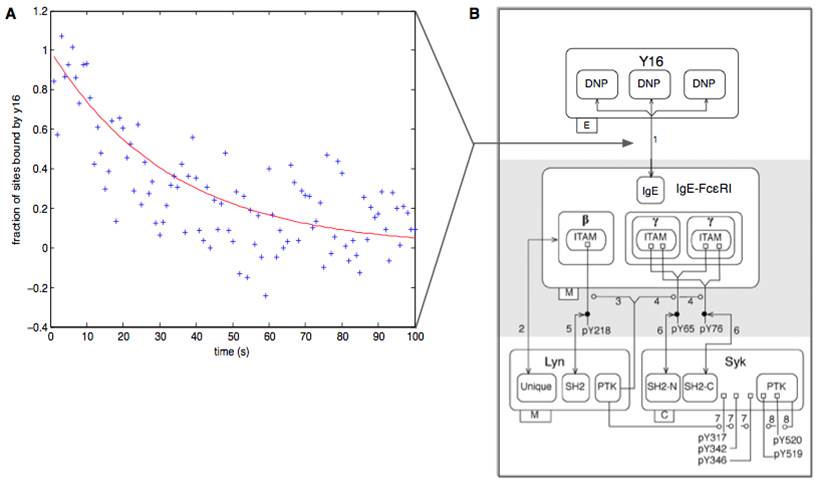
The role of ligand binding in FcεRI signal initiationInteractions of multivalent ligands with IgE-FcεRI can stimulate signaling in mast cells, but not all ligands give rise to the same outcomes. How does the structure of a ligand determine the response of a cell? To answer this question, we are investigating the binding properties and signaling capabilities of structurally defined ligands. Previous work has shown that increasing the distance between binding sites on a trivalent ligand can attenuate specific signaling events without affecting others (Sil et al., ACS Chem Biol, 2007), suggesting a branch point in the FcεRI signaling network. To better understand how these signals are initiated, we are characterizing equilibrium and kinetic properties of interactions using a number of approaches, including fluorescence quenching measurements that can be used to monitor occupancy of receptor sites. In addition, we are developing computational models that capture signaling kinetics in this system. These studies will help identify how differences in ligand characteristics can be translated into differences in receptor aggregation and cell signaling. Chylek, L.A., D.A. Holowka, B.A. Baird and W.S. Hlavacek: An interaction library for the FceRI signaling network. Front. Immunol. 5: 172 (2014). Chylek, L.A., D.A. Holowka, B.A. Baird, W.S. Hlavacek: Quantitative modeling of mast cell signaling. In Systems Immunology: An Introduction to Modeling Methods for Scientists (Jayajit Das, Ciriyam Jayaprakash, Editors), CRC Press, Boca Raton, FL, Ch. 13, pp. 213-226. |
|
 |